A general model for the bias in variant effect sizes due to population structure
Motivation
The effects of population structure in GWAS (genome-wide association studies) have been well-studied. It’s one of those problems that on a surface level is quite easy to grasp intuitively. And that sort of understanding is fine for most practical purposes. However, there’s still a lot of confusion around how population structure impacts/biases variant effect sizes, even among geneticists and particularly with respect to the difference between genetic and environmental stratification. I think some of this stems from a lack of good/easy/clear statistical descriptions of the effect of population stratification on GWAS. I worked this out while working on a recent paper 1 so I decided to post it in case others find it helpful.
The phenotype model
We begin by stating a generative model for some heritable phenotype
Here,
The estimated effect size of a variant
The ordinary least squares (OLS) effect size estimate of a predictor
In expectation,
Now, we know that
And the expected estimated effect size becomes:
Thus, the expected effect size estimate of a variant in a GWAS with population structure can be written as a sum of its own true effect size
Genetic stratification
Population structure induces long-range LD (linkage disequilibirum) between variants across the genome. This causes the estimated effect size of the test variant
In a panmictic population,
In other words, the bias due to background genetic effects can also be thought of as coming from the correlation between the test variant and the genotypic values (or true polygenic scores) in the population.
Environmental stratification
Any (known and unknown) environmental factor (
To illustrate, imagine a phenotype (let’s say height) which is influenced by the latitude at which individuals live such that people in the north tend to be taller and people in the south tend to be shorter (for whatever reason). Let’s denote the latitude of the
This is demonstrated in the figure below, which shows the true effect size on the x-axis and estimated effect size on the y-axis with colors representing the direction and magnitude of the correlation between frequency and latitude.
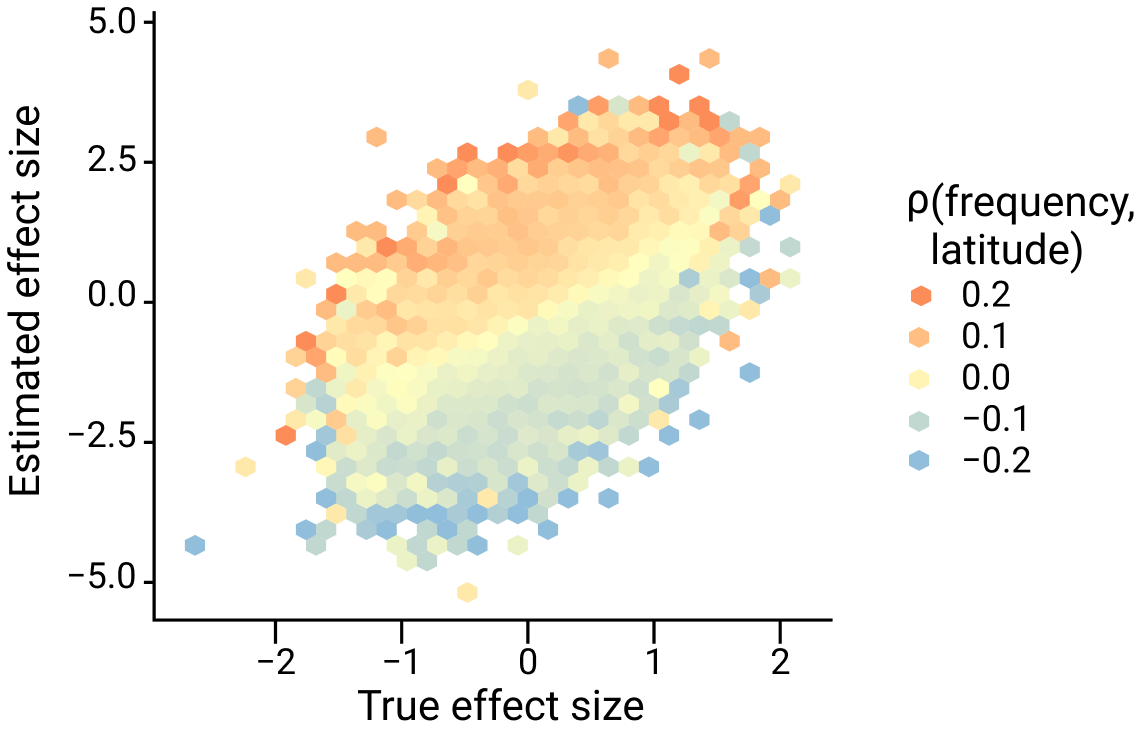
Figure adapted from Zaidi and Mathieson eLife 2020
Notice a few things:
The effect size of any variant whose frequency is correlated with latitude (i.e. for which
The effect size of alleles that are positively correlated with latitude (red colors) tends to be inflated (lies above the diagonal) whereas alleles that are negatively correlated with latitude (blue colors) tend to be biased downwards (below the diagonal).
The degree of this bias is greater for variants that are more strongly correlated with latitude (darker colors).
In summary, the estimated effect size of the
Technically, the allele frequency of a given variant could be correlated with an environmental effect (and therefore bias its effect size) even in unstructured/randomly mating populations (just by chance). But, this is very unlikely in an unstructured population. With more structure in the population, the distribution of allele frequencies in the population becomes less random, increasing the probability that they will be correlated with environmental effects. The LD between causal variants will increase as well (multi-locus Wahlund effect). Altogether, population structure will introduce bias in the effect size through both background genetic effects and environmental stratification.